Survival Analysis
Metabrick dataset is used below for demoing purposes.
The dataset can be found within examples/datasets/metabrick/ folder in the LogML repo. The configuration
file described below might be found in examples/configs/metabrick/survival_analysis.yaml file in the LogML repo.
Useful links:
Configuration
The following configuration file defines survival analysis for metabrick dataset:
1version: 0.2.6
2
3stratification:
4 - strata_id: cohort_1
5 query: 'cohort == 1.0'
6 - strata_id: cohort_2
7 query: 'cohort == 2.0'
8 - strata_id: cohort_3
9 query: 'cohort == 3.0'
10 - strata_id: cohort_4
11 query: 'cohort == 4.0'
12 - strata_id: cohort_5
13 query: 'cohort == 5.0'
14
15dataset_metadata:
16 modeling_specs:
17 # Overall survival setup, referenced below
18 OS:
19 time_column: overall_survival_months
20 event_column: overall_survival
21 # Interpretation of event column's values is study-specific,
22 # so explicit definition of 'uncensored' is required
23 event_query: 'overall_survival == 0'
24
25survival_analysis:
26 enable: True
27 problems:
28 # Reference to the OS modeling specification.
29 OS:
30 # Required transformations before proceeding to analyses.
31 dataset_preprocessing:
32 steps:
33 # Sample ID and stratification column are removed.
34 - transformer: drop_columns
35 params:
36 columns_to_include:
37 - patient_id
38 - cohort
39
40 methods:
41 - method_id: kaplan_meier
42 - method_id: optimal_cut_off
43 - method_id: cox
44
45report:
46 report_structure:
47 survival_analysis:
48 - OS
Stratification
stratification section defines a set of strata for which survival analysis will be run.
logml.configuration.stratification.Strata class defines each stratum.
In the example above there are 5 strata and each corresponds to some ‘cohort’ identifier.
Metadata definition
dataset_metadata/modeling_specs section defines all required survival setups (OS, PFS, etc.). For each
survival setup some reasonable alias should be introduced (OS - overall survival, PFS - progression free survival) and
then each alias is mapped to corresponding definition (logml.configuration.metadata.SurvivalTimeSpec).
In the example above there is the only one survival setup defined (using ‘OS’ alias) on top of the ‘overall_survival_month’ and ‘overall_survival’ columns.
Survival Analysis configuration
survival_analysis/problems section maps survival setup aliases to corresponding survival setup definitions
(logml.configuration.survival_analysis.SurvivalAnalysisSetup). Each survival setup specifies how dataset
should be preprocessed (dataset_preprocessing section) before applying target survival analysis
methods (methods section).
report/report_structure/survival_analysis section lists all survival setup aliases for which artifacts
should be included into the result report.
Univariate methods
As it was mentioned, before applying univariate methods a given dataset is preprocessed using the configured list of transformations.
Kaplan-Meier
The method consists of the following steps:
LogRank test is applied to all features (to what was left after preprocessing) - raw p-values are kept
for numerical features
medianthreshold is used to define ‘High’ and ‘Low’ groupscategorical features naturally define groupings for the test
FDR correction is applied to the result raw p-values
Features are ordered according to the corrected p-values
The method can be configured using logml.survival_analysis.extractors.kaplan_meier.KaplanMeierSAParams
class using params section.
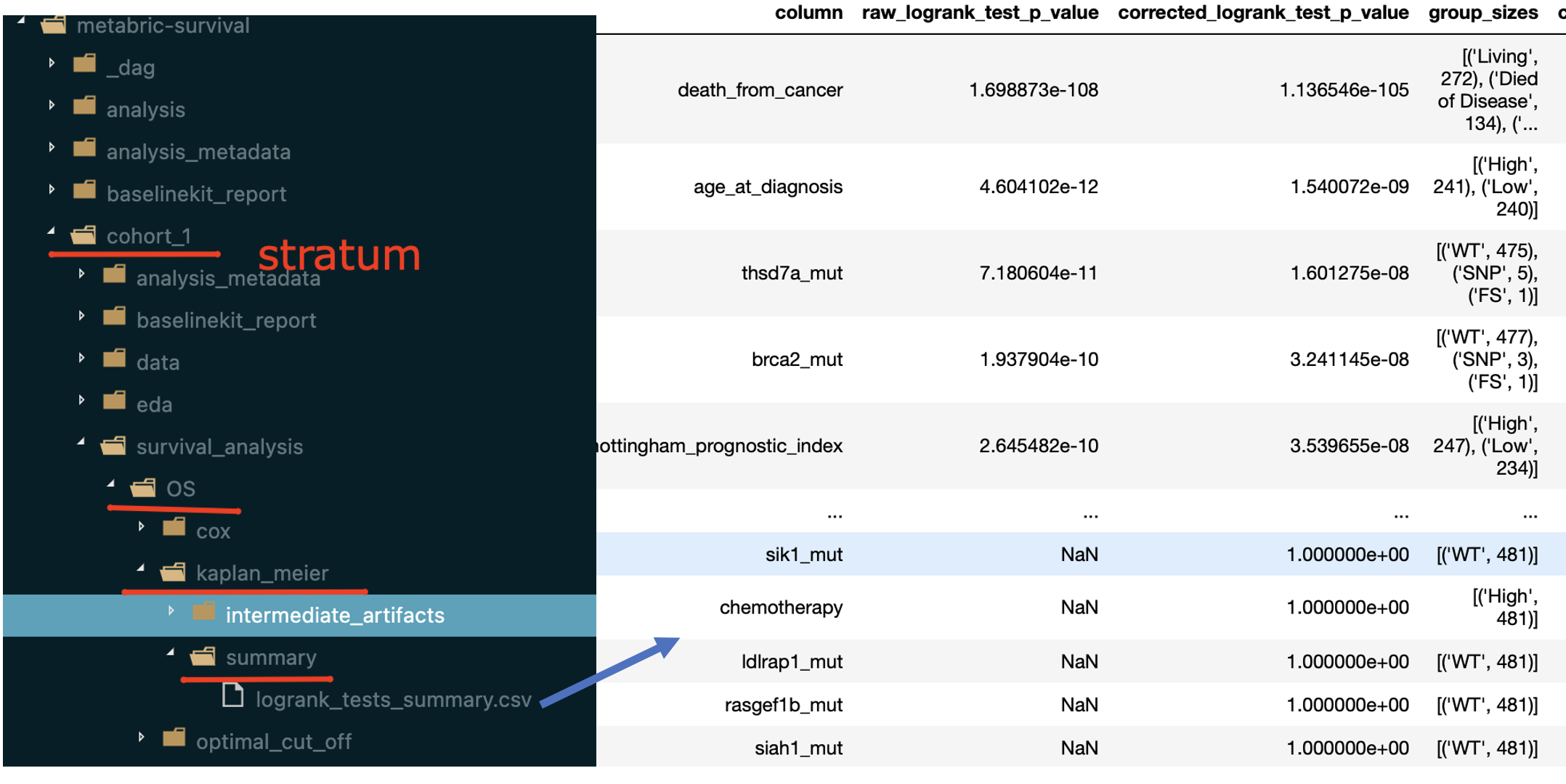
The result report contains the following artifacts:
table with features ranked by FDR-corrected p-values from LogRank tests
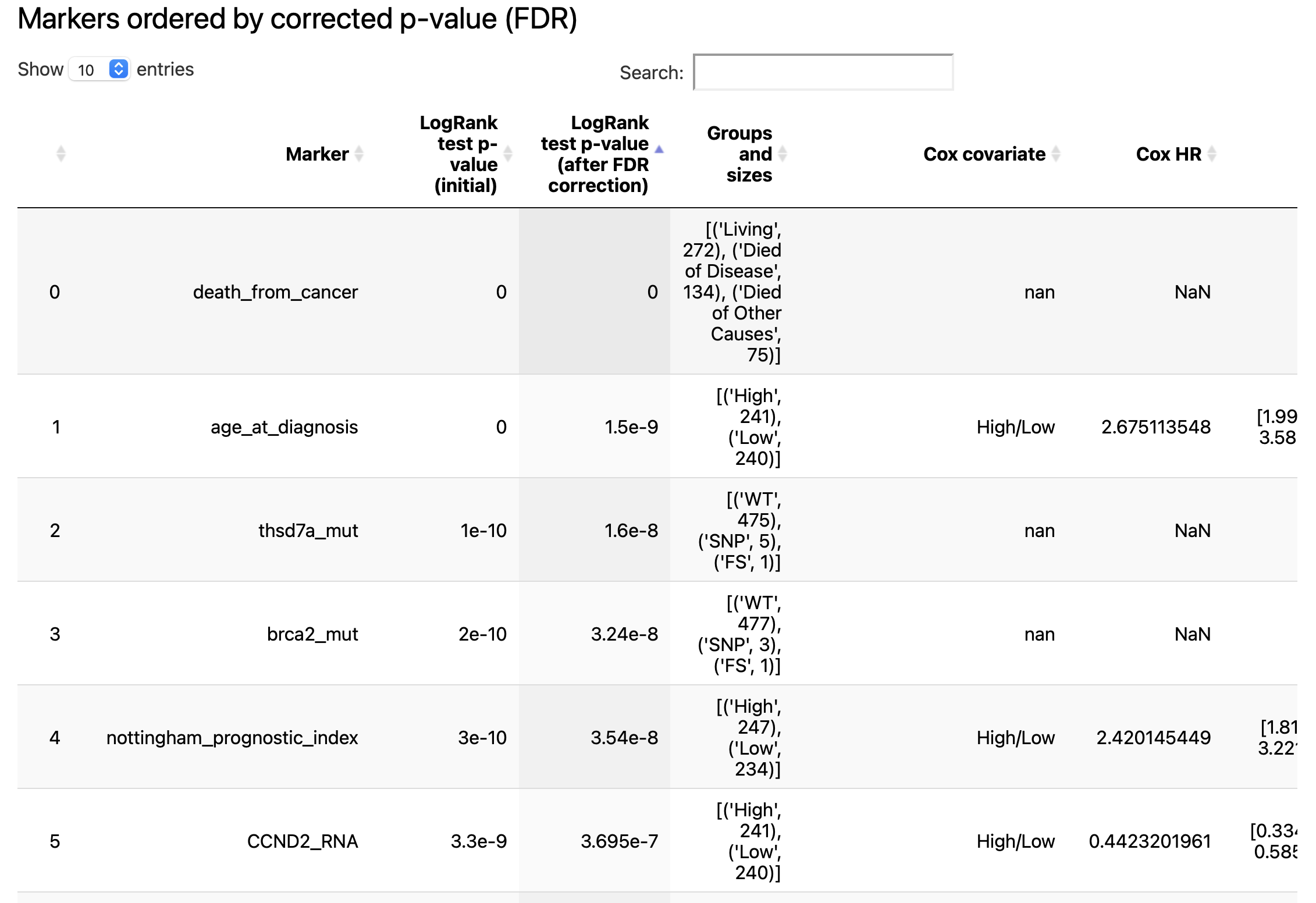
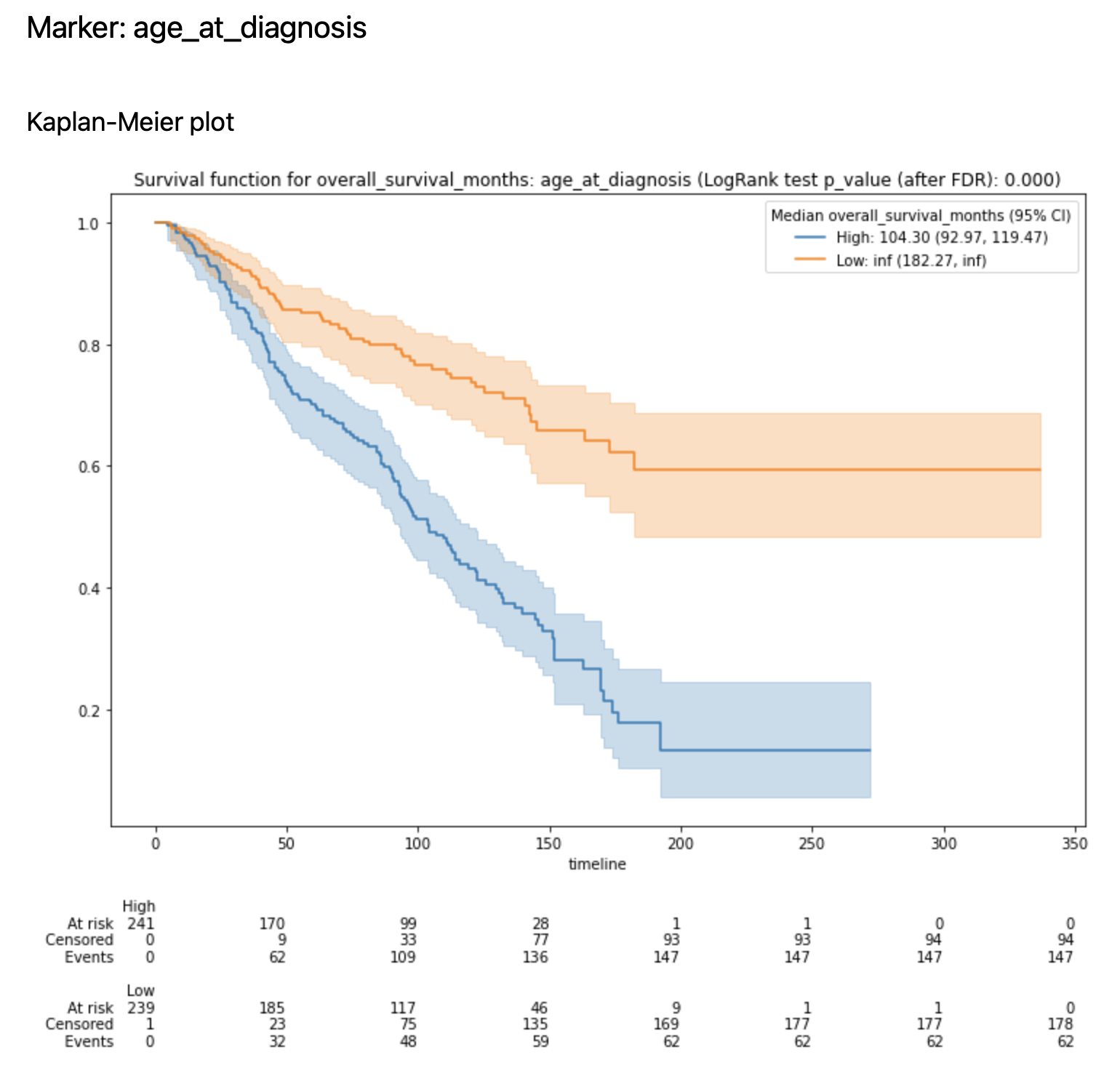
for a number of features (defined by
n_plots_to_showparameter) Kaplan-Meier plots are created
The summary table mentioned above can be found within dumped artifacts:
Optimal cut-off
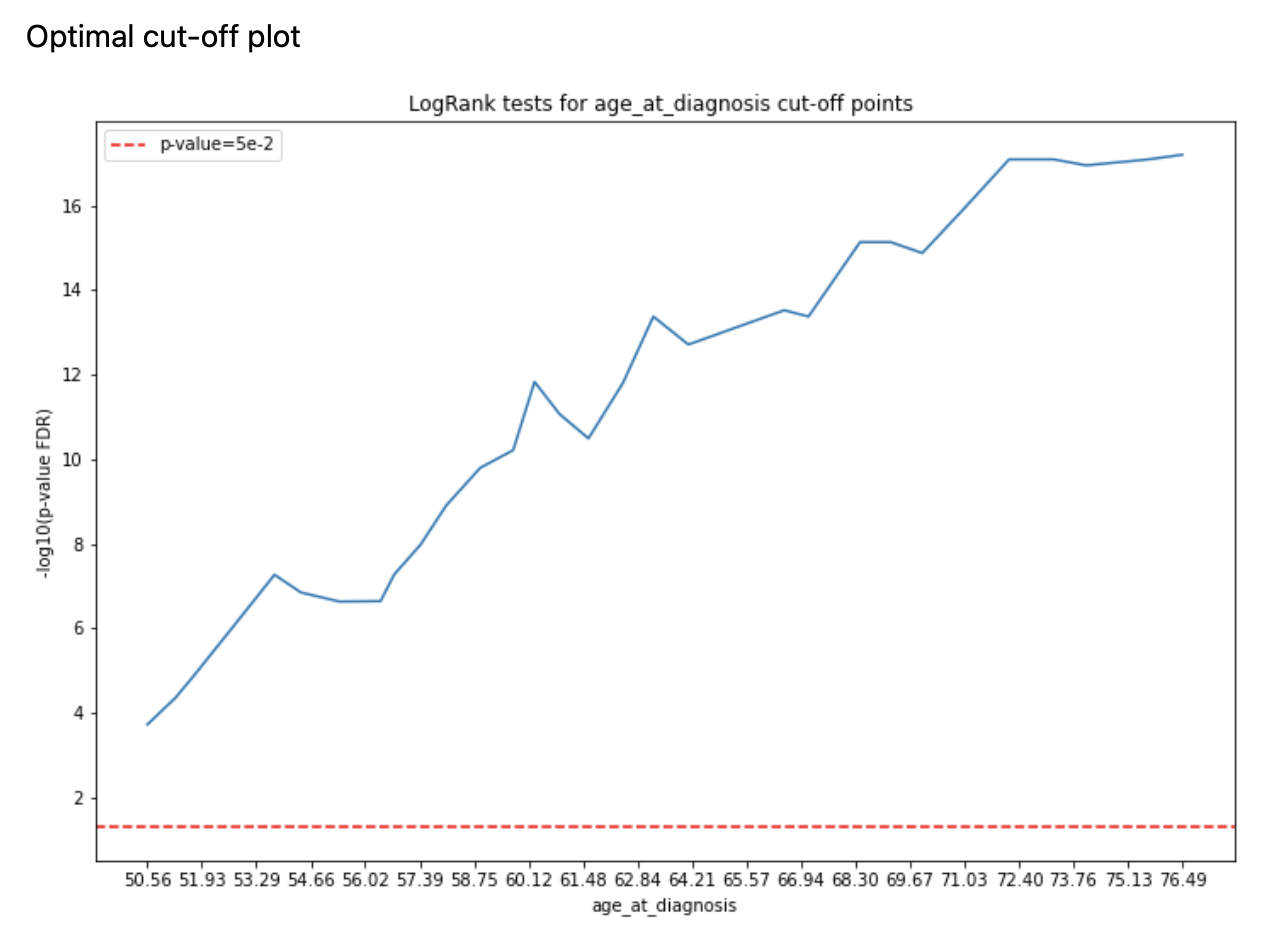
The method is based on the same schema as kaplan_meier method, the only difference is that for numerical features
we want to find a better threshold rather than median under the following constraints (defined via
logml.survival_analysis.extractors.optimal_cut_off.OptimalCutOffSAParams):
min_population- configures minimal fraction of samples within the smallest group (either ‘high’ or ‘low’), so that corner cases are avoided for which LogRank test returns unreasonable p-valuesn_percentiles- number of potential threshold to check
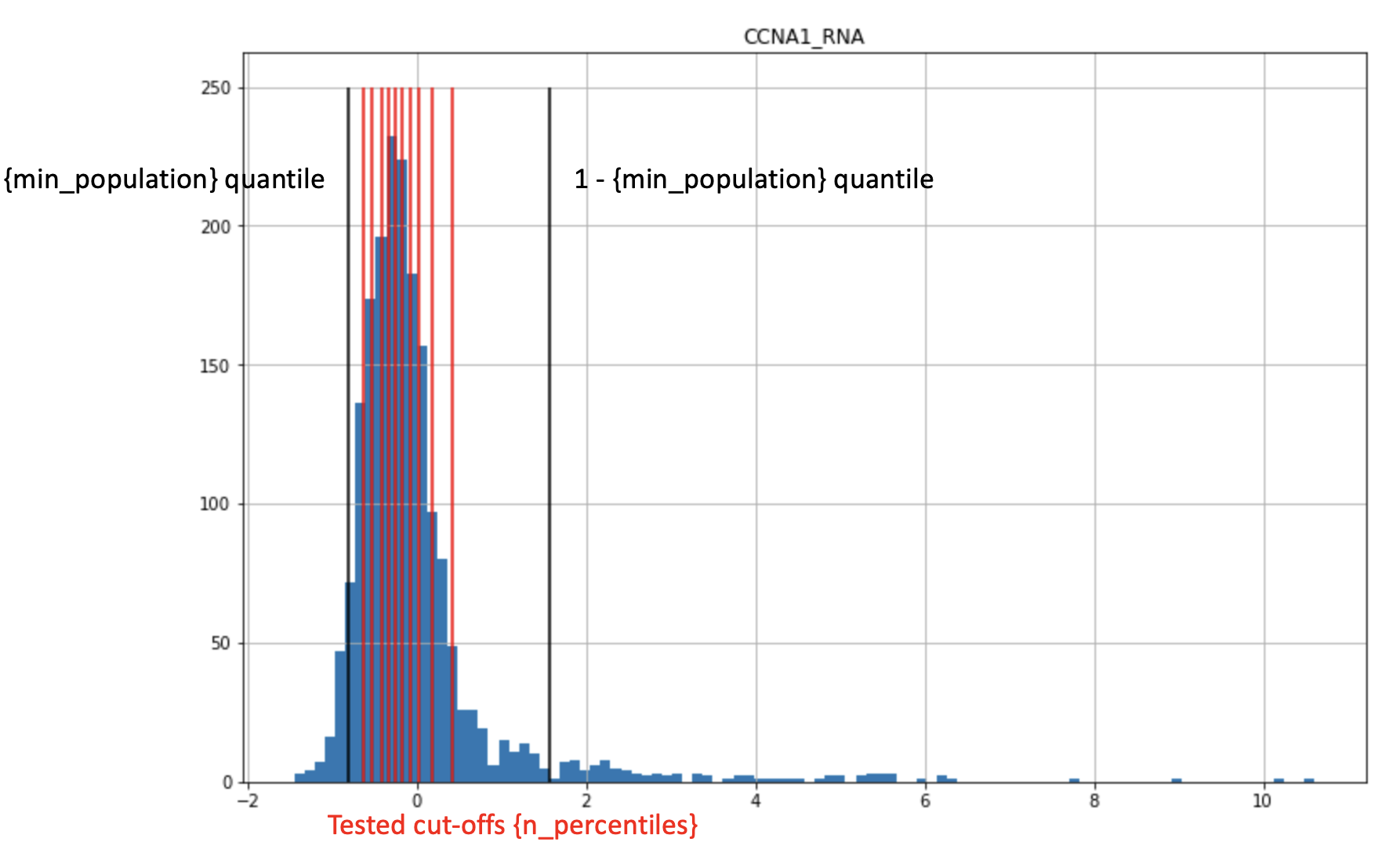
For all thresholds LogRank test is run and the best threshold is picked for a feature.
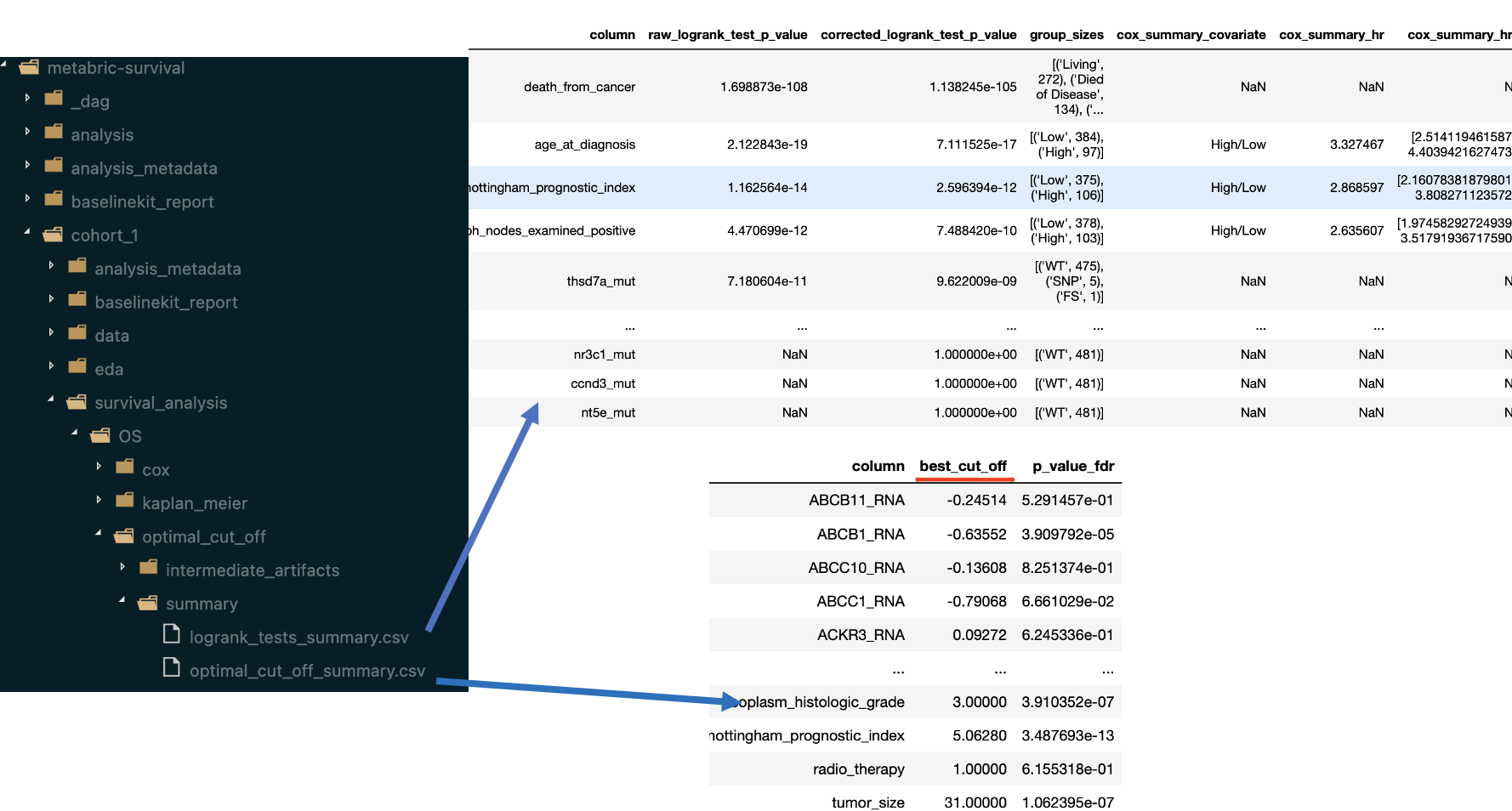
The result report contains the following artifacts:
table with features ranked by FDR-corrected p-values from LogRank tests
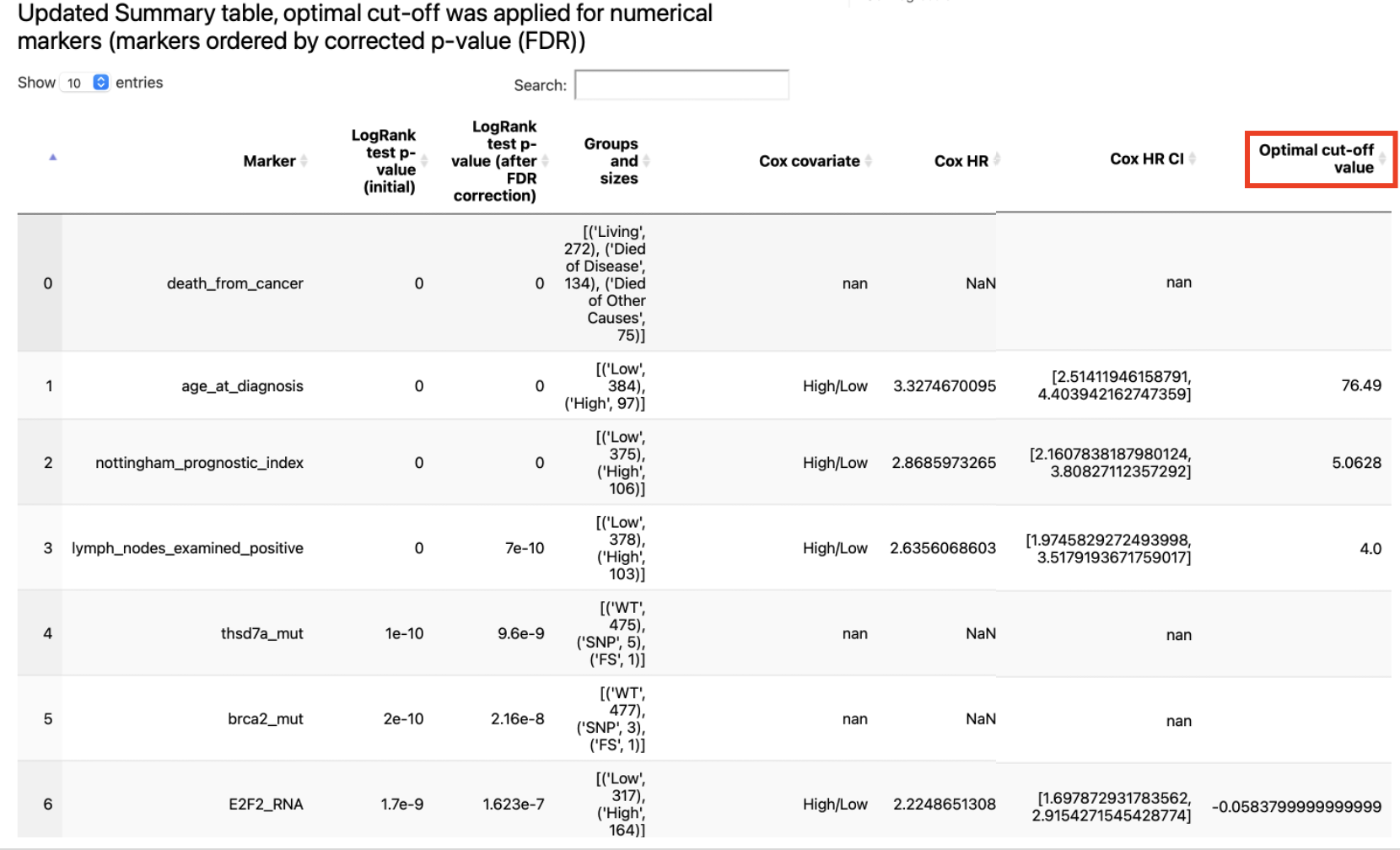
for a number of features (defined by
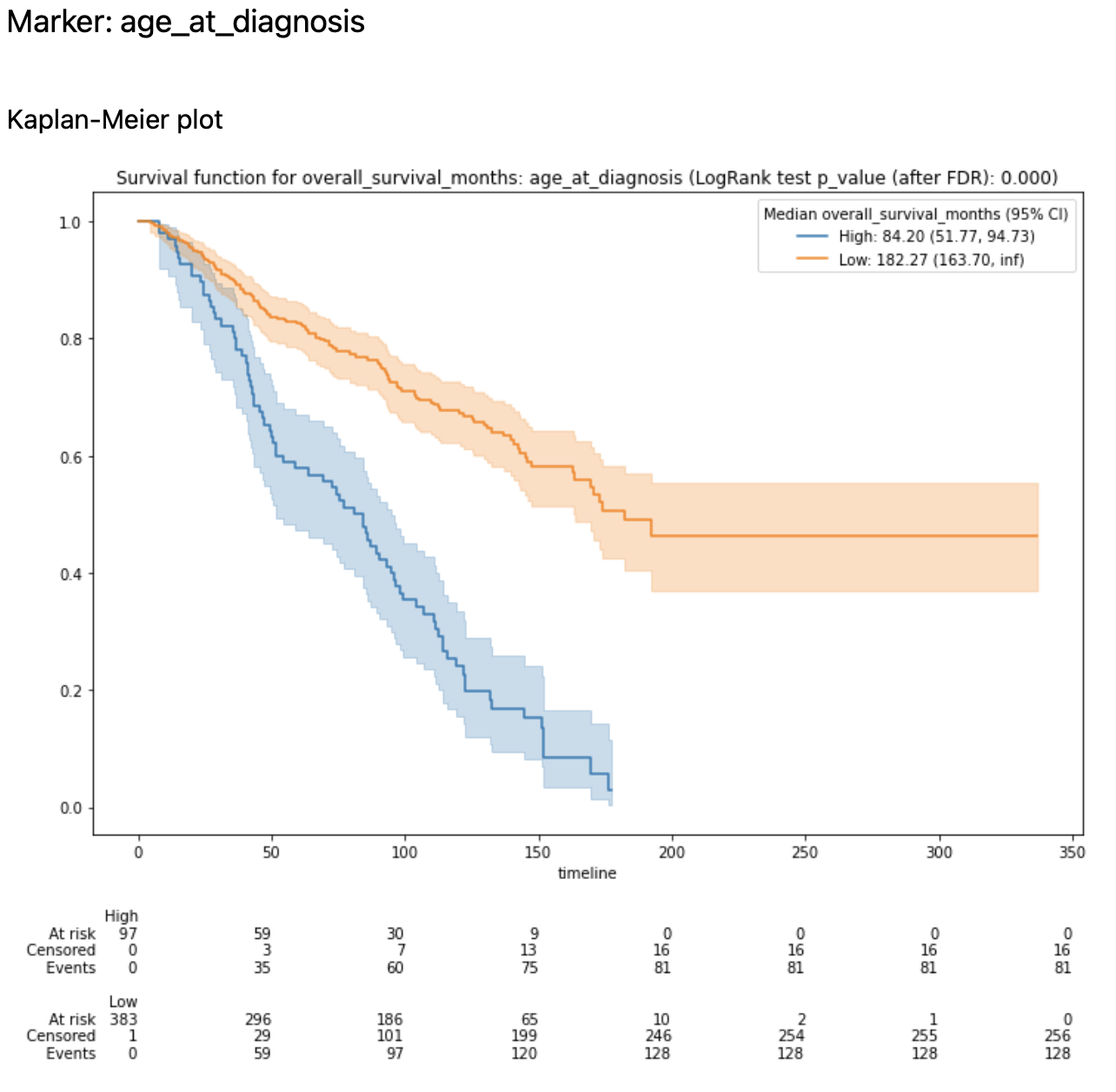
n_plots_to_showparameter) Kaplan-Meier plots are created (using optimal cut-offs) and Optimal Cut-Off plot is created (with LogRank test results for candidate thresholds)
The summary table mentioned above can be found within dumped artifacts:
Multivariate methods
Data Preprocessing
Before applying multivariate analysis methods a given dataset is prepared by adding a few additional transformations to the preprocessing pipeline that was defined via configuration file:
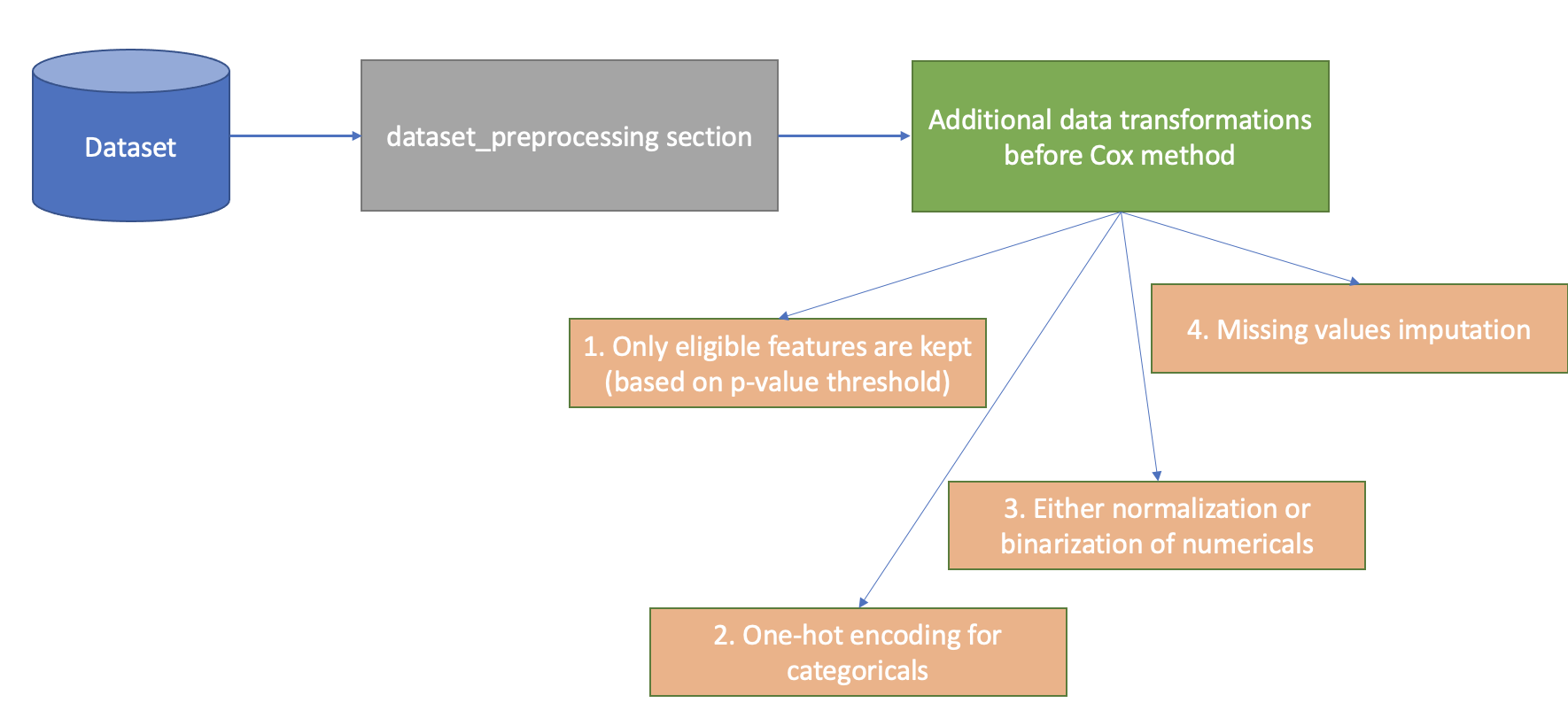
coxsurvival analysis method is strongly dependent on univariate methods, so the required prerequisite for running it isoptimal_cut_offmethod. Based on univariate summary table the target set of features is defines viaunivariate_p_value_thresholdparameter fromlogml.survival_analysis.extractors.cox.CoxSAParams- only features that are important enough in terms of univariate survival analysis (LogRank test p-value) are to be used in survival regression analysis (Cox).normalize_numericalsparameter defines whether numerical features should be normalized or based on the Optimal Cut-Off analysis results those features should be binarized (using best thresholds found).all categorical features are one-hot encoded
all missing values are imputed using Iterative Imputation (MICE)
Cox backward-forward method
The method is based on the following paper - Cox backward-forward feature selection. The idea is to come up with a set of features for which a Cox regression model contains only significant features. There are two phases:
‘backward’ - until there are insignificant features based on current Cox model, those features are removed and a new Cox model is created
‘forward’ - features that were removed are sequentially (in reverse order) are added to Cox model, and if it makes sense - those are kept
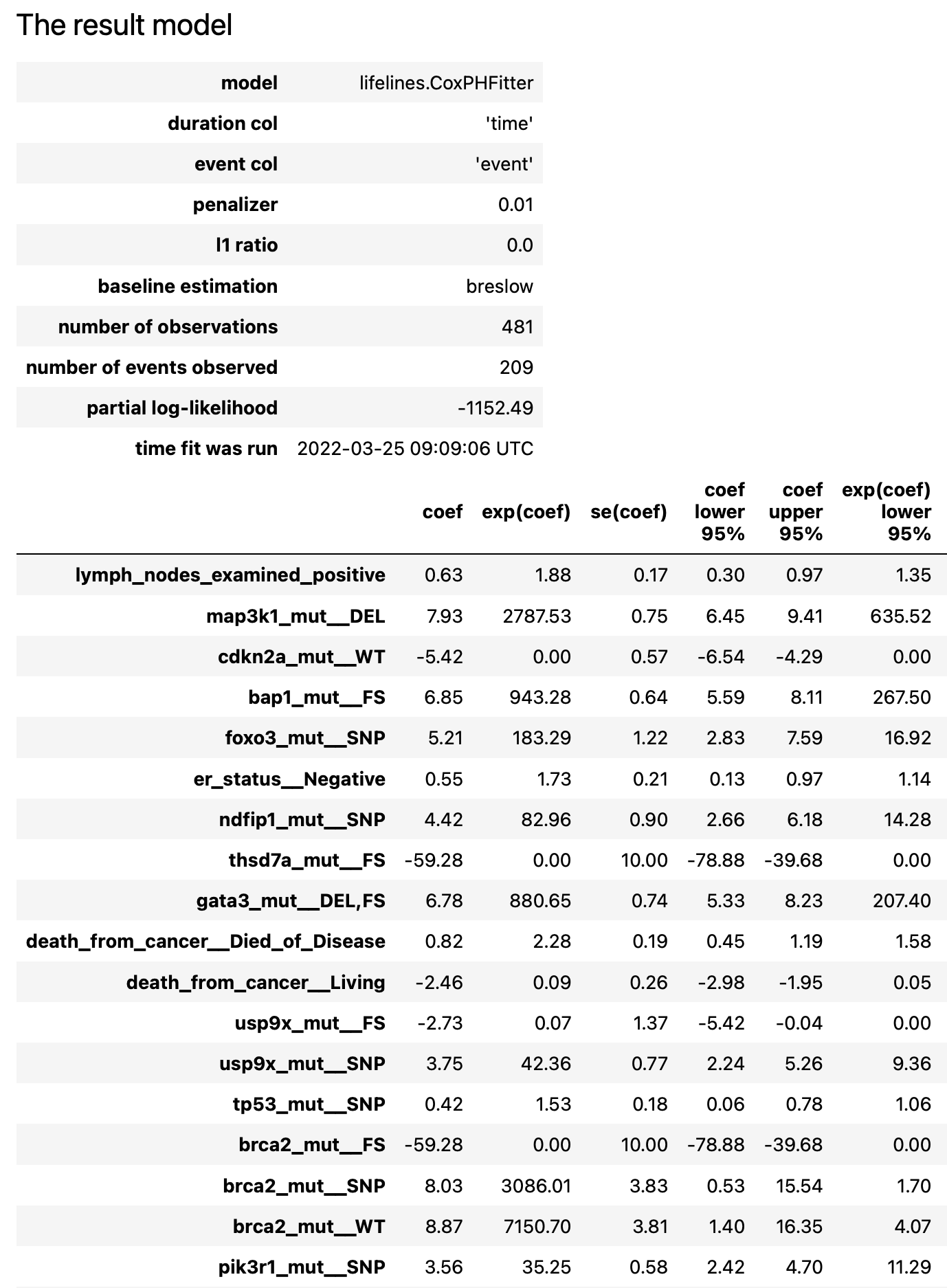
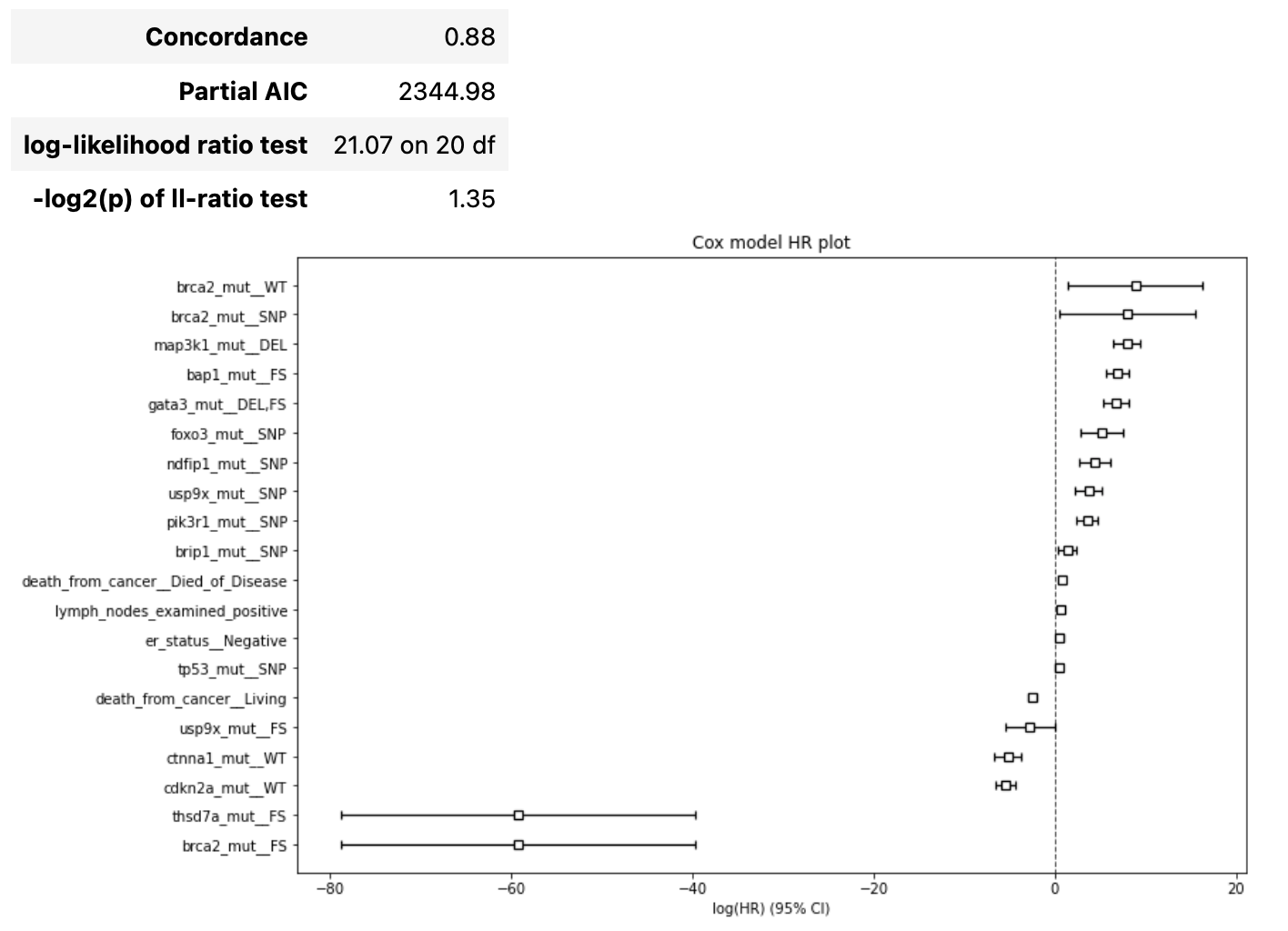
The result report contains the following artifacts:
initial Cox model summary
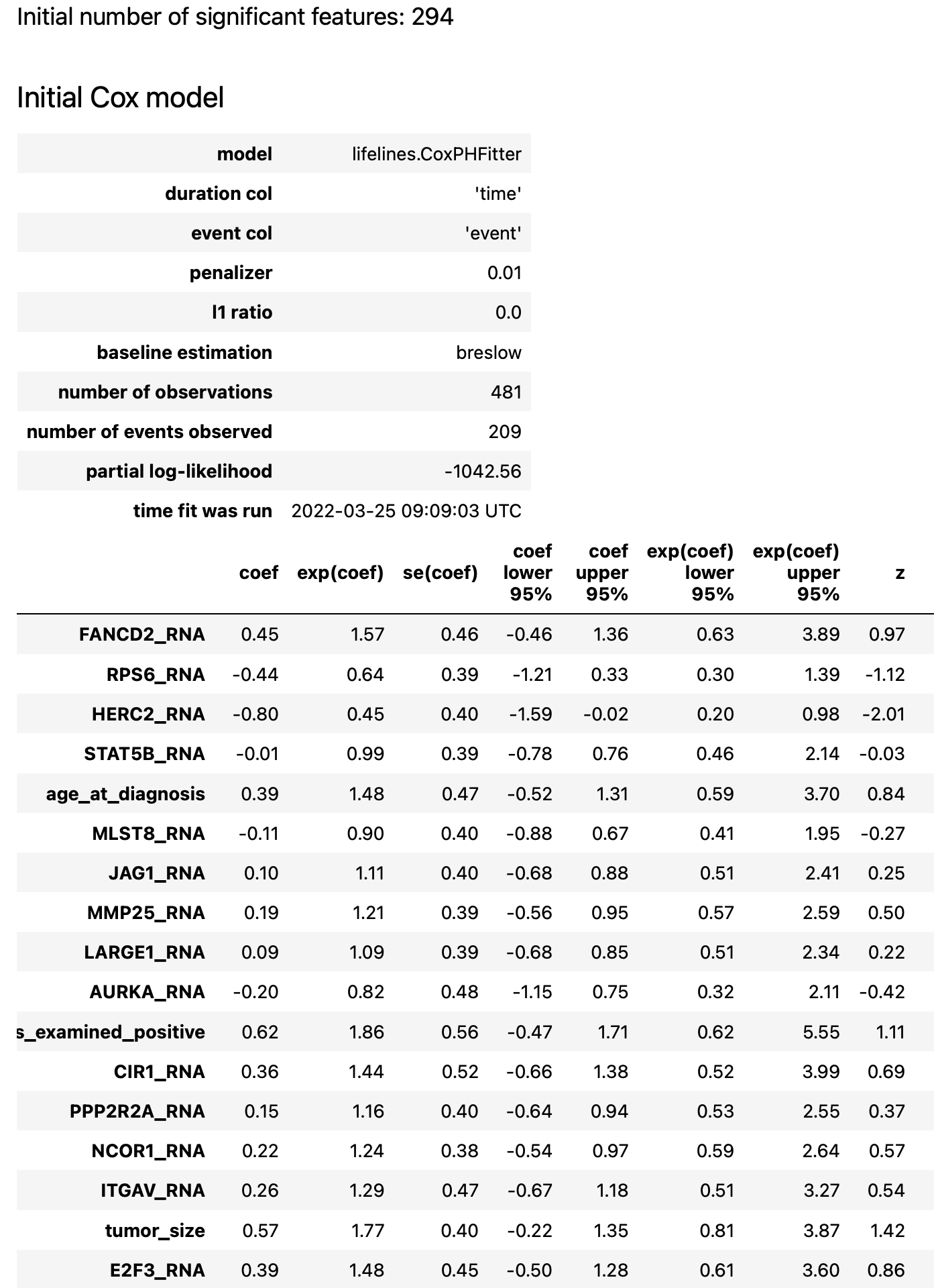
details about forward/backward phases


result Cox model summary